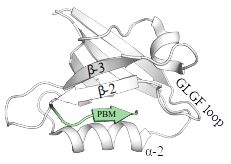
PDZ domains form a large protein family with ~270 members in the human proteome dispersed over ~150 proteins. Their name originates from 3 proteins where they were originally identified: PSD95/DLG1/ZO2. Their fold is highly conserved and consists of 5 to 6 β-strands that form two stacked antiparallel β-sheets and 2 α-helices. PDZ sequences most often contain a loop, bearing bearing the consensus "GLGF", located between the second and the third β-strands (β-2 and β-3). This loop serves for recognition of target motifs (see below).
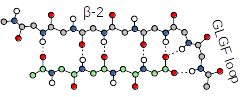
PDZ domains recognize short conserved PDZ-binding motifs (PBMs) mostly located at the extreme C-terminus of their target proteins.
There are approximately ~5000 putative in human. Therefore, at least 1/6th of the entire human proteome contains either, or both, a PDZ domain, or a putative PBM, building up a wide PDZ-PBM interactome that contributes to virtually all cellular processes and aspects of cellular life. PDZ-PBM interactions are relatively promiscuous. Typically, each binding motif is capable to detectably interact with dozens of PDZ domains with various affinities. Our database attempts to chart the affinity-based interactomic map of PDZ-PBM interactions exhaustively. The C-terminal carboxylate of PBMs is recognized by a loop, bearing the consensus "GLGF", located between the second and the third β-strands (β-2 and β-3) of the PDZ domains. The four last residues of the PBM, preceeding the carboxylate position, form a short beta-strand with the β-2 strand of the PDZ-fold.
PDZ-binding motifs are conventionally numbered backward from the C-terminal residue (position 0). The sequences of C-terminal PBMs fall into three main classes (Luck et al., 2012). The last C-terminal residue (position 0) is almost always hydrophobic (mainly, Leu/Val/Ile). The third last residue (position -2) can be Ser/Thr (class 1), Val/Tyr/Phe (class 2), or Asp/Glu (class 3). A PDZ ligand peptide adopts a short antiparallel β-sheet conformation in contact with the β-2 strand of the PDZ domain. This way, their interaction is mediated by several main chain directed interactions. The antepenultimate residue (position -2) is exposed to the α-2 helix of the PDZ domain that contains a critical residue for interaction. This residue is very often a His which can make a hydrogen bond with a complementary Ser/Thr residue of a class 1 PBM, or a hydrophobic residue which can make interaction of hydrophobic (class 2) of acidic (class 3) PBMs. Based on these interactions, we can sort class 1/2/3 PBMs using the consensus motifs [ST].[ACVILF]$, [VLIFY].[ACVILF]$,[DE].[ACVILF]$, respectively.
One must note that this classification scheme was made a priori to the exploration of the PDZ-PBM interactome and according to our current knowledge we can observe that the existence of a class-specific acceptor residue on the PDZ domain is not always necessary for strong interaction. In addition, these artificial consensus motifs can lead to the underestimation of the real number in the human PBMome. Our current database already contains a few example motifs that show binding activity towards multiple PDZ-domains despite they do not match with the conventional consensus motif of PBMs. Besides, while these consensus motifs are limited to only the last 3 residues of C-terminal sequences, residues outside the last 5 residues can also contribute to the PDZ binding via transient, fuzzy interactions. Finally, a number of "internal" PBMs, not situated at the C-terminal extremity of proteins, have also been described.
Based on the consensus motif found in the ELM database, PBMs are defined as [STVLIFYDE].[ACVILFM]$ (where . and $ denotes any residue and the C-terminus, respectively). A more restrictive consensus [STVYFED].[LVI]$ can be also used to find some of the most reliable motifs. Using this later definition in SLiMSearch4, one can find
~2000 PBMs in the human proteome, including ~1000 class 1 and ~500-500 class 2 and class 3 motifs. Using the loose consensus motif, the approximate number of PBMs is ~4000. Since each PBM can potentially interact with the 266 human PDZ domains, the complete landscape of the PDZ-PBM interactome covers more than a million domain-motif (4000x266) and >600000 (4000x150) protein-protein potential interactions. Several PBMs are also subjected to site-specific post-translational modifications creating an even larger and more diverse set of putative PBMs. In reality, putative motifs that includes C-terminal PBMs with less common consensus motifs and internal motifs may greatly increase the estimated number of the human PBMome, greatly increasing the size of the PDZ-PBM domain-motif interactome.
Download the current database from here in csv format
- The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains K Luck et al. FEBS Letters, 2012