Details of experiments between Q9Y4H2 and P63104
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: IRS2
Protein A fragment: 574-583
Protein A site: 14-3-3_pThr579
Protein A modification: (TPO)579
Protein A sequence: RTYSLTTPAR
Protein A construct: RTYSL(pThr)TPAR (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): <1
Immobilized partner concentration in holdup experiment (10-6M): 132
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 1
Measured pKd: 6
Experiment series 2
Protein A protein: IRS2
Protein A fragment: 718-727
Protein A site: 14-3-3_pSer723
Protein A modification: (SEP)723
Protein A sequence: RTFPASGGGY
Protein A construct: RTFPA(pSer)GGGY (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.31
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 6.84
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.53
Experiment series 3
Protein A protein: IRS2
Protein A fragment: 427-436
Protein A site: 14-3-3_pSer432
Protein A modification: (SEP)432
Protein A sequence: HSRSMSMPVA
Protein A construct: HSRSM(pSer)MPVA (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.88
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 6.72
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.72
Experiment series 4
Protein A protein: IRS2
Protein A fragment: 379-388
Protein A site: 14-3-3_pSer384
Protein A modification: (SEP)384
Protein A sequence: GARPVSVAGS
Protein A construct: GARPV(pSer)VAGS (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.27
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 5.98
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.44
Experiment series 5
Protein A protein: IRS2
Protein A fragment: 603-612
Protein A site: 14-3-3_pSer608
Protein A modification: (SEP)608
Protein A sequence: ATFSGSAGRL
Protein A construct: ATFSG(pSer)AGRL (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.17
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.9
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 6
Protein A protein: IRS2
Protein A fragment: 1171-1180
Protein A site: 14-3-3_pSer1176
Protein A modification: (SEP)1176
Protein A sequence: RHNSASVENV
Protein A construct: RHNSA(pSer)VENV (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.15
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.33
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: IRS2
Protein A fragment: 725-734
Protein A site: 14-3-3_pSer730
Protein A modification: (SEP)730
Protein A sequence: GGYKASSPAE
Protein A construct: GGYKA(pSer)SPAE (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.05
Holdup BI standard deviation: 0.12
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.42
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 8
Protein A protein: IRS2
Protein A fragment: 518-527
Protein A site: 14-3-3_pSer523
Protein A modification: (SEP)523
Protein A sequence: SNTPESIAET
Protein A construct: SNTPE(pSer)IAET (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.06
Holdup BI standard deviation: 0.21
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.31
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: IRS2
Protein A fragment: 341-350
Protein A site: 14-3-3_pSer346
Protein A modification: (SEP)346
Protein A sequence: RSRTDSLAAT
Protein A construct: RSRTD(pSer)LAAT (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.02
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.23
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 10
Protein A protein: IRS2
Protein A fragment: 1159-1168
Protein A site: 14-3-3_pSer1164
Protein A modification: (SEP)1164
Protein A sequence: TPVSPSFAHN
Protein A construct: TPVSP(pSer)FAHN (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.01
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.16
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 11
Protein A protein: IRS2
Protein A fragment: 1278-1287
Protein A site: 14-3-3_pSer1283
Protein A modification: (SEP)1283
Protein A sequence: PGDKSSWGRT
Protein A construct: PGDKS(pSer)WGRT (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0
Holdup BI standard deviation: 0.09
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.04
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 12
Protein A protein: IRS2
Protein A fragment: 574-583
Protein A site: 14-3-3_pThr579
Protein A modification: (TPO)579
Protein A sequence: RTYSLTTPAR
Protein A construct: biotin-ttds-RTYSL(pThr)TPAR (HPLC-purified synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): 6.3
Standard deviation of dissociation constant (10-6M): 0.4
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit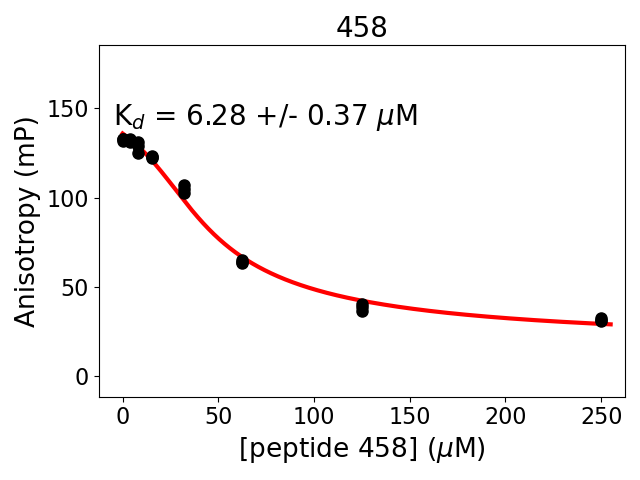
Date of measurement: 2022.01.19
Number of measurements: 3
Measured pKd: 5.2
Experiment series 13
Protein A protein: IRS2
Protein A fragment: 574-583
Protein A site: 14-3-3_pThr579
Protein A modification: (TPO)579
Protein A sequence: RTYSLTTPAR
Protein A construct: biotin-ttds-RTYSL(pThr)TPAR (HPLC-purified synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): 4.6
Standard deviation of dissociation constant (10-6M): 0.5
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit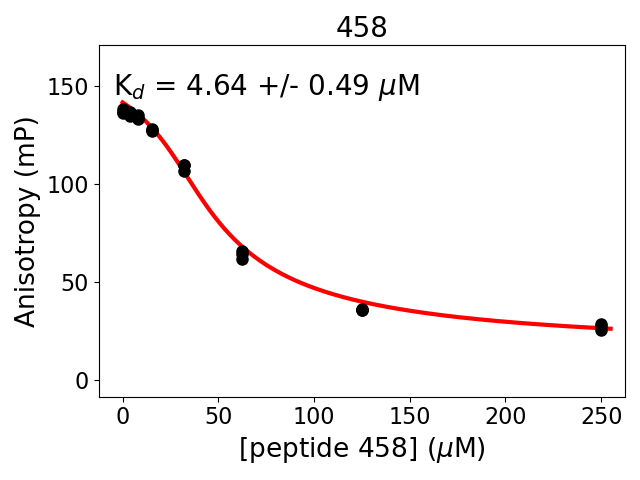
Date of measurement: 2022.02.10
Number of measurements: 3
Measured pKd: 5.33