Details of experiments between Q9P2M7 and P31946
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: CGN
Protein A fragment: 126-135
Protein A site: 14-3-3_pSer131
Protein A modification: (SEP)131
Protein A sequence: LLRSHSQASL
Protein A construct: LLRSH(pSer)QASL (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: 0.33
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 134
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 5.71
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 3.57
Experiment series 2
Protein A protein: CGN
Protein A fragment: 144-153
Protein A site: 14-3-3_pSer149
Protein A modification: (SEP)149
Protein A sequence: SNRSNSMLEL
Protein A construct: SNRSN(pSer)MLEL (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: 0.41
Holdup BI standard deviation: 0.07
Immobilized partner concentration in holdup experiment (10-6M): 102
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.99
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.84
Experiment series 3
Protein A protein: CGN
Protein A fragment: 126-135
Protein A site: 14-3-3_pSer131
Protein A modification: (SEP)131
Protein A sequence: LLRSHSQASL
Protein A construct: LLRSH(pSer)QASL (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: 0.28
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 102
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.59
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.59
Experiment series 4
Protein A protein: CGN
Protein A fragment: 144-153
Protein A site: 14-3-3_pSer149
Protein A modification: (SEP)149
Protein A sequence: SNRSNSMLEL
Protein A construct: SNRSN(pSer)MLEL (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: 0.66
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 134
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.16
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 4.15
Experiment series 5
Protein A protein: CGN
Protein A fragment: 66-75
Protein A site: 14-3-3_pSer71
Protein A modification: (SEP)71
Protein A sequence: EKGGDSFGVQ
Protein A construct: EKGGD(pSer)FGVQ (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: -0.07
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 102
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.99
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 6
Protein A protein: CGN
Protein A fragment: 66-75
Protein A site: 14-3-3_pSer71
Protein A modification: (SEP)71
Protein A sequence: EKGGDSFGVQ
Protein A construct: EKGGD(pSer)FGVQ (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: -0.06
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 134
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.65
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: CGN
Protein A fragment: 327-336
Protein A site: 14-3-3_pSer332
Protein A modification: (SEP)332
Protein A sequence: VRRKVSLVLE
Protein A construct: VRRKV(pSer)LVLE (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: 0.05
Holdup BI standard deviation: 0.69
Immobilized partner concentration in holdup experiment (10-6M): 134
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.05
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 8
Protein A protein: CGN
Protein A fragment: 327-336
Protein A site: 14-3-3_pSer332
Protein A modification: (SEP)332
Protein A sequence: VRRKVSLVLE
Protein A construct: VRRKV(pSer)LVLE (crude synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Average holdup BI: -0.01
Holdup BI standard deviation: 0.17
Immobilized partner concentration in holdup experiment (10-6M): 102
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.04
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: CGN
Protein A fragment: 126-135
Protein A site: 14-3-3_pSer131
Protein A modification: (SEP)131
Protein A sequence: LLRSHSQASL
Protein A construct: biotin-ttds-LLRSH(pSer)QASL (HPLC-purified synthetic peptide)
Protein B protein: YWHAB
Protein B fragment: 1-246
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3beta
Measured dissociation constant (10-6M): 90
Standard deviation of dissociation constant (10-6M): 20
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit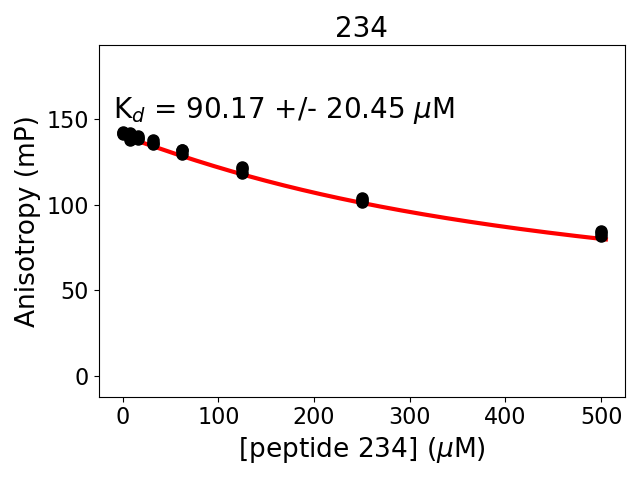
Date of measurement: 2022.02.08
Number of measurements: 3
Measured pKd: 4.05