Details of experiments between the fragments Q6UVM3~1126-1135 and P78352~306-403
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: KCNT2
Protein A fragment: 1126-1135
Protein A site: PBM
Protein A sequence: GQDSREETQL
Protein A construct: biotin-ttds-GQDSREETQL
Protein B protein: DLG4
Protein B fragment: 306-403
Protein B site: PDZ_3
Protein B construct: his6-MBP-TEVsite-PDZ
Measured dissociation constant (10-6M): 240.56
Standard deviation of dissociation constant (10-6M): 64.17
Experiment method: competitive FP
Details of affinity fitting: Competitive binding equation in ProFit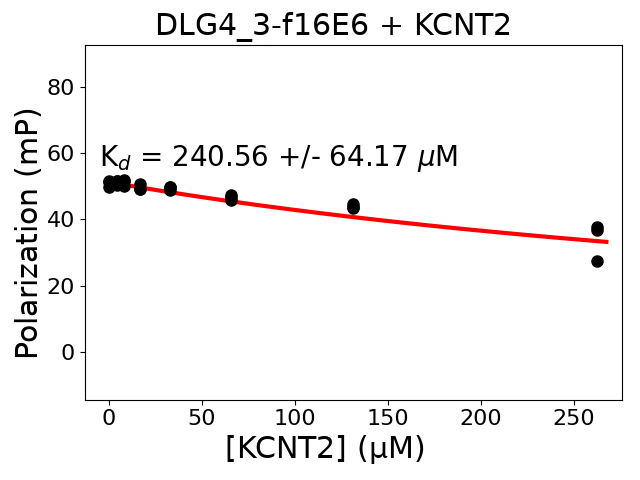
PUBMED ID of publication: 36115835
Number of measurements: 3
Measured pKd: 3.62
Experiment series 2
Protein A protein: KCNT2
Protein A fragment: 1126-1135
Protein A site: PBM
Protein A sequence: GQDSREETQL
Protein A construct: biotin-ttds-GQDSREETQL
Protein B protein: DLG4
Protein B fragment: 306-403
Protein B site: PDZ_3
Protein B construct: his6-MBP-TEVsite-PDZ
Average holdup BI: -0.03
Holdup BI standard deviation: 0.02
Normalized holdup BI: -0.04
Normalized holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 13.3
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
PUBMED ID of publication: 36115835
Number of measurements: 2
The affinity was below the detection threshold of the assay.