Details of experiments between Q5VZ89 and P63104
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: DENND4C
Protein A fragment: 1657-1666
Protein A site: 14-3-3_pSer1662
Protein A modification: (SEP)1662
Protein A sequence: GISTVSLPNS
Protein A construct: GISTV(pSer)LPNS (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.72
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 8.42
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.28
Experiment series 2
Protein A protein: DENND4C
Protein A fragment: 1635-1644
Protein A site: 14-3-3_pSer1640
Protein A modification: (SEP)1640
Protein A sequence: LTRSHSVGGP
Protein A construct: LTRSH(pSer)VGGP (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.64
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 8.04
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.12
Experiment series 3
Protein A protein: DENND4C
Protein A fragment: 1320-1329
Protein A site: 14-3-3_pSer1325
Protein A modification: (SEP)1325
Protein A sequence: KERSTSLSAL
Protein A construct: KERST(pSer)LSAL (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.77
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.25
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.39
Experiment series 4
Protein A protein: DENND4C
Protein A fragment: 1618-1627
Protein A site: 14-3-3_pSer1623
Protein A modification: (SEP)1623
Protein A sequence: TMKISSVPNS
Protein A construct: TMKIS(pSer)VPNS (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.42
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.12
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.74
Experiment series 5
Protein A protein: DENND4C
Protein A fragment: 1273-1282
Protein A site: 14-3-3_pSer1278
Protein A modification: (SEP)1278
Protein A sequence: LERRSSLPLD
Protein A construct: LERRS(pSer)LPLD (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.38
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 5.94
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.67
Experiment series 6
Protein A protein: DENND4C
Protein A fragment: 1094-1103
Protein A site: 14-3-3_pSer1099
Protein A modification: (SEP)1099
Protein A sequence: IARTHSFENV
Protein A construct: IARTH(pSer)FENV (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.09
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.07
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: DENND4C
Protein A fragment: 1179-1188
Protein A site: 14-3-3_pSer1184
Protein A modification: (SEP)1184
Protein A sequence: RNKRSSLYGI
Protein A construct: RNKRS(pSer)LYGI (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.28
Holdup BI standard deviation: 0.07
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 3.89
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.48
Experiment series 8
Protein A protein: DENND4C
Protein A fragment: 984-993
Protein A site: 14-3-3_pSer989
Protein A modification: (SEP)989
Protein A sequence: IVKVPSGIFD
Protein A construct: IVKVP(pSer)GIFD (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.36
Holdup BI standard deviation: 0.35
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.77
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: DENND4C
Protein A fragment: 948-957
Protein A site: 14-3-3_pSer953
Protein A modification: (SEP)953
Protein A sequence: LIRLESIDNH
Protein A construct: LIRLE(pSer)IDNH (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.08
Holdup BI standard deviation: 0.09
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.13
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 10
Protein A protein: DENND4C
Protein A fragment: 1341-1350
Protein A site: 14-3-3_pSer1346
Protein A modification: (SEP)1346
Protein A sequence: VVNSLSGLKL
Protein A construct: VVNSL(pSer)GLKL (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.11
Holdup BI standard deviation: 0.23
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.55
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 11
Protein A protein: DENND4C
Protein A fragment: 960-969
Protein A site: 14-3-3_pSer965
Protein A modification: (SEP)965
Protein A sequence: IVAKHSQPSP
Protein A construct: IVAKH(pSer)QPSP (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.04
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.3
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 12
Protein A protein: DENND4C
Protein A fragment: 1661-1670
Protein A site: 14-3-3_pSer1666
Protein A modification: (SEP)1666
Protein A sequence: VSLPNSLQEV
Protein A construct: VSLPN(pSer)LQEV (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.02
Holdup BI standard deviation: 0.17
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.11
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 13
Protein A protein: DENND4C
Protein A fragment: 1320-1329
Protein A site: 14-3-3_pSer1325
Protein A modification: (SEP)1325
Protein A sequence: KERSTSLSAL
Protein A construct: biotin-ttds-KERST(pSer)LSAL (HPLC-purified synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): 64
Standard deviation of dissociation constant (10-6M): 13
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit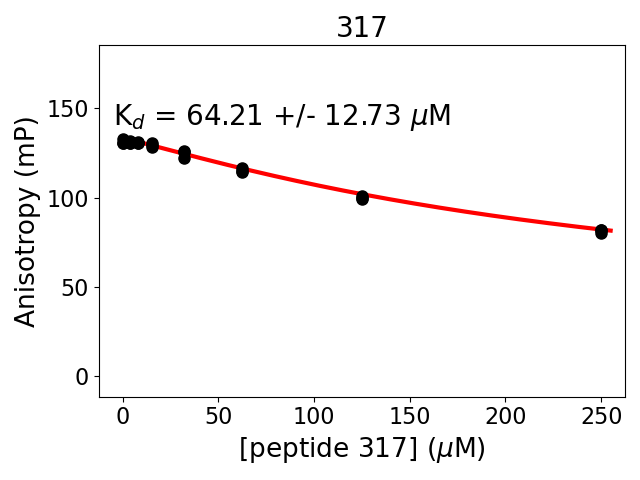
Date of measurement: 2022.01.19
Number of measurements: 3
Measured pKd: 4.19
Experiment series 14
Protein A protein: DENND4C
Protein A fragment: 1320-1329
Protein A site: 14-3-3_pSer1325
Protein A modification: (SEP)1325
Protein A sequence: KERSTSLSAL
Protein A construct: biotin-ttds-KERST(pSer)LSAL (HPLC-purified synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): 58
Standard deviation of dissociation constant (10-6M): 0
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit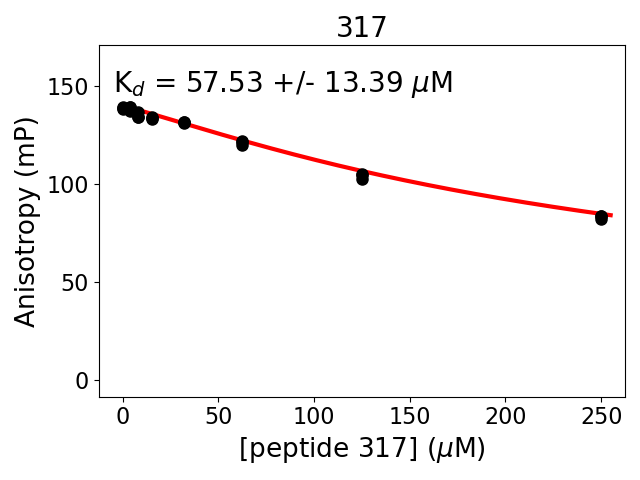
Date of measurement: 2022.02.10
Number of measurements: 3
Measured pKd: 4.24