Details of experiments between the fragments Q5T2W1~2-107 and Q7Z628~587-596
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: NHERF3
Protein A fragment: 2-107
Protein A site: PDZ_1
Protein A construct: his6-MBP-TEVsite-PDZ
Protein B protein: NET1
Protein B fragment: 587-596
Protein B site: PBM
Protein B sequence: SGGKRKETLV
Protein B construct: biotin-ttds-SGGKRKETLV
Average holdup BI: 0.47
Immobilized partner concentration in holdup experiment (10-6M): 35
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
PUBMED ID of publication: 36115835
Number of measurements: 1
Measured pKd: 4.42
Experiment series 2
Protein A protein: NHERF3
Protein A fragment: 2-107
Protein A site: PDZ_1
Protein A construct: his6-MBP-TEVsite-PDZ
Protein B protein: NET1
Protein B fragment: 587-596
Protein B site: PBM
Protein B sequence: SGGKRKETLV
Protein B construct: biotin-ttds-SGGKRKETLV
Measured dissociation constant (10-6M): 73.04
Standard deviation of dissociation constant (10-6M): 26.23
Experiment method: competitive FP
Details of affinity fitting: Competitive binding equation in ProFit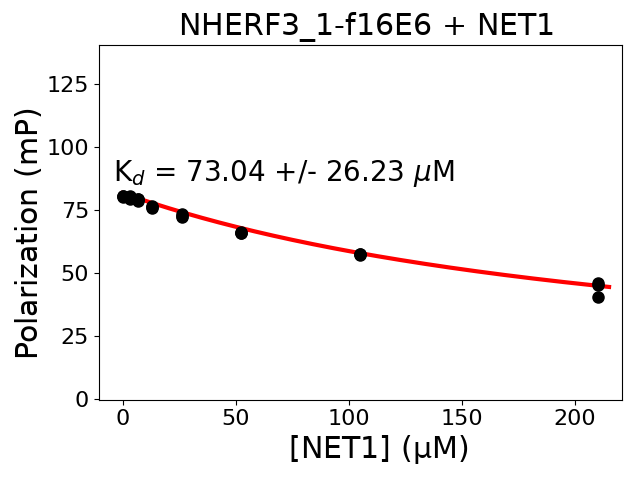
PUBMED ID of publication: 36115835
Number of measurements: 3
Measured pKd: 4.14
Experiment series 3
Protein A protein: NHERF3
Protein A fragment: 2-107
Protein A site: PDZ_1
Protein A construct: his6-MBP-TEVsite-PDZ
Protein B protein: NET1
Protein B fragment: 587-596
Protein B site: PBM
Protein B sequence: SGGKRKETLV
Protein B construct: biotin-ttds-SGGKRKETLV
Average holdup BI: 0.26
Holdup BI standard deviation: 0.01
Normalized holdup BI: 0.27
Normalized holdup BI standard deviation: 0.01
Immobilized partner concentration in holdup experiment (10-6M): 17.9
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
PUBMED ID of publication: 36115835
Number of measurements: 2
Measured pKd: 4.35
Experiment series 4
Protein A protein: NHERF3
Protein A fragment: 2-107
Protein A site: PDZ_1
Protein A construct: his6-MBP-TEVsite-PDZ
Protein B protein: NET1
Protein B fragment: 587-596
Protein B site: PBM
Protein B sequence: SGGKRKETLV
Protein B construct: biotin-ttds-SGGKRKETLV
Average holdup BI: 0.21
Normalized holdup BI: 0.23
Immobilized partner concentration in holdup experiment (10-6M): 17.9
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Experimental details: not reported internal standards of holdup layout #5
Number of measurements: 1
Measured pKd: 4.25
Experiment series 5
Protein A protein: NHERF3
Protein A fragment: 2-107
Protein A site: PDZ_1
Protein A construct: his6-MBP-TEVsite-PDZ
Protein B protein: NET1
Protein B fragment: 587-596
Protein B site: PBM
Protein B sequence: SGGKRKETLV
Protein B construct: Biotin-ado-ado-SGGKRKETLV
Average holdup BI: 0.41
Normalized holdup BI: 0.44
Immobilized partner concentration in holdup experiment (10-6M): 18
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Experimental details: Crude peptide synthesis product. The peptide concentration in the affinity conversion step may differ from reality. Not reported internal standards of holdup layout #6
PUBMED ID of publication: 36115835
Number of measurements: 1
Measured pKd: 4.68