Details of experiments between Q15311 and Q96L92
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: RALBP1
Protein A fragment: 646-655
Protein A site: PBM
Protein A sequence: PSRDRKETSI
Protein A construct: biotin-ttds-PSRDRKETSI
Protein B protein: SNX27
Protein B fragment: 40-141
Protein B site: PDZ
Protein B construct: his6-MBP-TEVsite-PDZ
Measured dissociation constant (10-6M): 71.14
Standard deviation of dissociation constant (10-6M): 11.23
Experiment method: competitive FP
Details of affinity fitting: Competitive binding equation in ProFit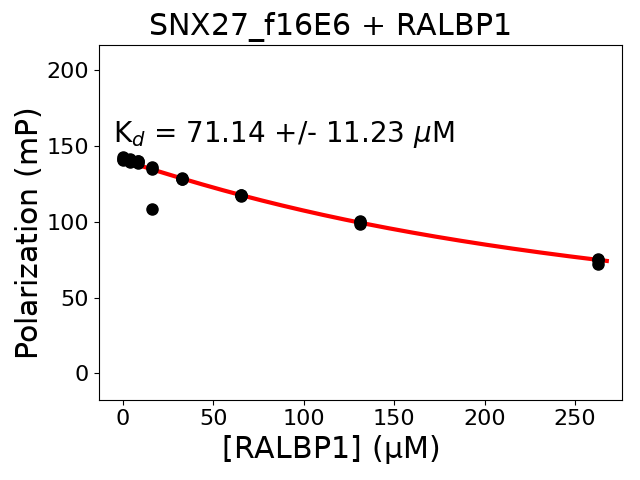
PUBMED ID of publication: 36115835
Number of measurements: 3
Measured pKd: 4.15
Experiment series 2
Protein A protein: RALBP1
Protein A fragment: 646-655
Protein A site: PBM
Protein A sequence: PSRDRKETSI
Protein A construct: biotin-ttds-PSRDRKETSI
Protein B protein: SNX27
Protein B fragment: 40-141
Protein B site: PDZ
Protein B construct: his6-MBP-TEVsite-PDZ
Average holdup BI: 0.4
Immobilized partner concentration in holdup experiment (10-6M): 20
Experiment method: SAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normal layout
PUBMED ID of publication: 36115835
Number of measurements: 1
Measured pKd: 4.55
Experiment series 3
Protein A protein: RALBP1
Protein A fragment: 646-655
Protein A site: PBM
Protein A sequence: PSRDRKETSI
Protein A construct: biotin-ttds-PSRDRKETSI
Protein B protein: SNX27
Protein B fragment: 40-141
Protein B site: PDZ
Protein B construct: his6-MBP-TEVsite-PDZ
Average holdup BI: 0.31
Holdup BI standard deviation: 0.03
Normalized holdup BI: 0.31
Normalized holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 28
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
PUBMED ID of publication: 36115835
Number of measurements: 3
Measured pKd: 4.23
Experiment series 4
Protein A protein: RALBP1
Protein A fragment: 646-655
Protein A site: PBM
Protein A sequence: PSRDRKETSI
Protein A construct: biotin-ttds-PSRDRKETSI
Protein B protein: SNX27
Protein B fragment: 40-141
Protein B site: PDZ
Protein B construct: his6-MBP-TEVsite-PDZ
Average holdup BI: 0.34
Normalized holdup BI: 0.34
Immobilized partner concentration in holdup experiment (10-6M): 18
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Experimental details: not reported internal standards of holdup layout #6
PUBMED ID of publication: 36115835
Number of measurements: 1
Measured pKd: 4.49
Experiment series 5
Protein A protein: RALBP1
Protein A fragment: 1-655
Protein A construct: endogenous protein (Jurkat)
Protein B protein: SNX27
Protein B fragment: 40-141
Protein B site: PDZ
Protein B construct: avi(biotin)-his6-MBP-TEVsite-PDZ
Average holdup BI: 0.5
Immobilized partner concentration in holdup experiment (10-6M): 10
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.81
Experiment method: nHU_Jurkat
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.08.03.
Experimental details: -log10(P Two-sided unpaired T-test (6 vs 3))
PUBMED ID of publication: 36542723
Number of measurements: 3
The affinity was below the detection threshold of the assay.