Details of experiments between P63104 and Q6R327
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1214-1223
Protein B site: 14-3-3_pSer1219
Protein B modification: (SEP)1219
Protein B sequence: KIRSQSFNTD
Protein B construct: KIRSQ(pSer)FNTD (crude synthetic peptide)
Average holdup BI: 0.51
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.96
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.9
Experiment series 2
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1279-1288
Protein B site: 14-3-3_pSer1284
Protein B modification: (SEP)1284
Protein B sequence: LSKSNSVSLV
Protein B construct: LSKSN(pSer)VSLV (crude synthetic peptide)
Average holdup BI: 0.41
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.54
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.72
Experiment series 3
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1297-1306
Protein B site: 14-3-3_pSer1302
Protein B modification: (SEP)1302
Protein B sequence: PRRAQSLKAP
Protein B construct: PRRAQ(pSer)LKAP (crude synthetic peptide)
Average holdup BI: 0.77
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 5.66
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.41
Experiment series 4
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1128-1137
Protein B site: 14-3-3_pThr1133
Protein B modification: (TPO)1133
Protein B sequence: NRRIRTLTEP
Protein B construct: NRRIR(pThr)LTEP (crude synthetic peptide)
Average holdup BI: 0.34
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.62
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.58
Experiment series 5
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1380-1389
Protein B site: 14-3-3_pSer1385
Protein B modification: (SEP)1385
Protein B sequence: FMKALSYASL
Protein B construct: FMKAL(pSer)YASL (crude synthetic peptide)
Average holdup BI: 0.56
Holdup BI standard deviation: 0.16
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.16
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.98
Experiment series 6
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1130-1139
Protein B site: 14-3-3_pThr1135
Protein B modification: (TPO)1135
Protein B sequence: RIRTLTEPSV
Protein B construct: RIRTL(pThr)EPSV (crude synthetic peptide)
Average holdup BI: 0.22
Holdup BI standard deviation: 0.13
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.75
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1079-1088
Protein B site: 14-3-3_pSer1084
Protein B modification: (SEP)1084
Protein B sequence: IKDKNSFPFF
Protein B construct: IKDKN(pSer)FPFF (crude synthetic peptide)
Average holdup BI: 0.44
Holdup BI standard deviation: 0.19
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.72
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.77
Experiment series 8
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1172-1181
Protein B site: 14-3-3_pSer1177
Protein B modification: (SEP)1177
Protein B sequence: TGSTPSIGEN
Protein B construct: TGSTP(pSer)IGEN (crude synthetic peptide)
Average holdup BI: -0.14
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.55
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1383-1392
Protein B site: 14-3-3_pSer1388
Protein B modification: (SEP)1388
Protein B sequence: ALSYASLDKE
Protein B construct: ALSYA(pSer)LDKE (crude synthetic peptide)
Average holdup BI: 0.04
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.46
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 10
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1327-1336
Protein B site: 14-3-3_pThr1332
Protein B modification: (TPO)1332
Protein B sequence: AFGYATLKRL
Protein B construct: AFGYA(pThr)LKRL (crude synthetic peptide)
Average holdup BI: -0.11
Holdup BI standard deviation: 0.29
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.43
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 11
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1133-1142
Protein B site: 14-3-3_pSer1138
Protein B modification: (SEP)1138
Protein B sequence: TLTEPSVDFN
Protein B construct: TLTEP(pSer)VDFN (crude synthetic peptide)
Average holdup BI: -0.02
Holdup BI standard deviation: 0.1
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.12
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 12
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 378-387
Protein B site: 14-3-3_pSer383
Protein B modification: (SEP)383
Protein B sequence: PHRARSRPDL
Protein B construct: PHRAR(pSer)RPDL (crude synthetic peptide)
Average holdup BI: 0.04
Holdup BI standard deviation: 0.3
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.09
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 13
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1214-1223
Protein B site: 14-3-3_pSer1219
Protein B modification: (SEP)1219
Protein B sequence: KIRSQSFNTD
Protein B construct: biotin-ttds-KIRSQ(pSer)FNTD (HPLC-purified synthetic peptide)
Measured dissociation constant (10-6M): 163
Standard deviation of dissociation constant (10-6M): 33
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit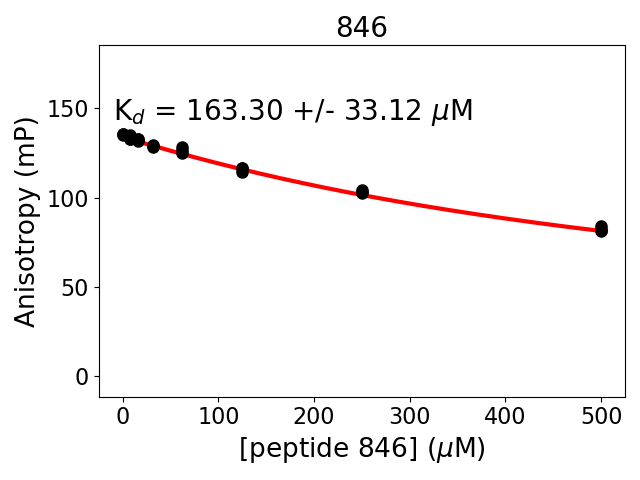
Date of measurement: 2022.01.19
Number of measurements: 3
Measured pKd: 3.79
Experiment series 14
Protein A protein: YWHAZ
Protein A fragment: 1-245
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3zeta
Protein B protein: RICTOR
Protein B fragment: 1214-1223
Protein B site: 14-3-3_pSer1219
Protein B modification: (SEP)1219
Protein B sequence: KIRSQSFNTD
Protein B construct: biotin-ttds-KIRSQ(pSer)FNTD (HPLC-purified synthetic peptide)
Measured dissociation constant (10-6M): 114
Standard deviation of dissociation constant (10-6M): 21
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit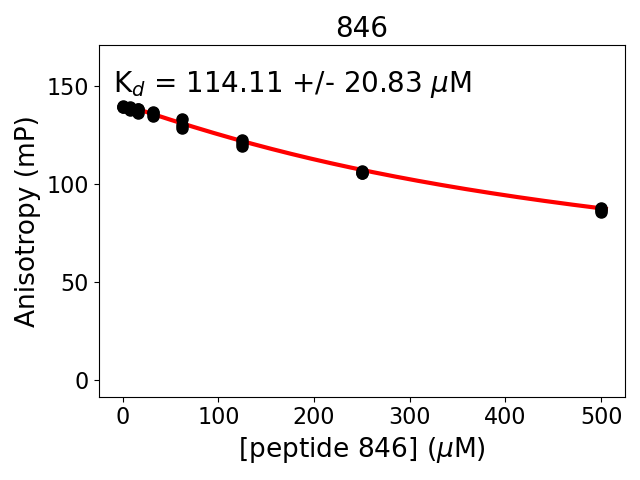
Date of measurement: 2022.02.10
Number of measurements: 3
Measured pKd: 3.94