Details of experiments between P62258 and Q6IQ23
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 533-542
Protein B site: 14-3-3_pThr538
Protein B modification: (TPO)538
Protein B sequence: RHGSPTAPI-
Protein B construct: FRHGSP(pThr)API (crude synthetic peptide)
Measured dissociation constant (10-6M): <10
Immobilized partner concentration in holdup experiment (10-6M): 143
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 1
Measured pKd: 5
Experiment series 2
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 443-452
Protein B site: 14-3-3_pSer448
Protein B modification: (SEP)448
Protein B sequence: KGDSRSLPLD
Protein B construct: KGDSR(pSer)LPLD (crude synthetic peptide)
Average holdup BI: 0.64
Holdup BI standard deviation: 0.01
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 9.03
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 4.18
Experiment series 3
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1008-1017
Protein B site: 14-3-3_pThr1013
Protein B modification: (TPO)1013
Protein B sequence: ESRYQTLPGR
Protein B construct: ESRYQ(pThr)LPGR (crude synthetic peptide)
Average holdup BI: 0.64
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.74
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 4.18
Experiment series 4
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 443-452
Protein B site: 14-3-3_pSer448
Protein B modification: (SEP)448
Protein B sequence: KGDSRSLPLD
Protein B construct: KGDSR(pSer)LPLD (crude synthetic peptide)
Average holdup BI: 0.64
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.61
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.1
Experiment series 5
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1008-1017
Protein B site: 14-3-3_pThr1013
Protein B modification: (TPO)1013
Protein B sequence: ESRYQTLPGR
Protein B construct: ESRYQ(pThr)LPGR (crude synthetic peptide)
Average holdup BI: 0.66
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 6.97
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.14
Experiment series 6
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 560-569
Protein B site: 14-3-3_pSer565
Protein B modification: (SEP)565
Protein B sequence: VPRSISVPPS
Protein B construct: VPRSI(pSer)VPPS (crude synthetic peptide)
Average holdup BI: 0.73
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 5.25
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.28
Experiment series 7
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 981-990
Protein B site: 14-3-3_pSer986
Protein B modification: (SEP)986
Protein B sequence: LRSYVSEPEL
Protein B construct: LRSYV(pSer)EPEL (crude synthetic peptide)
Average holdup BI: 0.61
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.73
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.05
Experiment series 8
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 981-990
Protein B site: 14-3-3_pSer986
Protein B modification: (SEP)986
Protein B sequence: LRSYVSEPEL
Protein B construct: LRSYV(pSer)EPEL (crude synthetic peptide)
Average holdup BI: 0.47
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.42
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 3.89
Experiment series 9
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1032-1041
Protein B site: 14-3-3_pThr1037
Protein B modification: (TPO)1037
Protein B sequence: IAPYVTLRRG
Protein B construct: IAPYV(pThr)LRRG (crude synthetic peptide)
Average holdup BI: 0.33
Holdup BI standard deviation: 0.07
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.41
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.55
Experiment series 10
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 467-476
Protein B site: 14-3-3_pThr472
Protein B modification: (TPO)472
Protein B sequence: PENYQTLPKS
Protein B construct: PENYQ(pThr)LPKS (crude synthetic peptide)
Average holdup BI: 0.12
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.4
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 11
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 467-476
Protein B site: 14-3-3_pThr472
Protein B modification: (TPO)472
Protein B sequence: PENYQTLPKS
Protein B construct: PENYQ(pThr)LPKS (crude synthetic peptide)
Average holdup BI: 0.17
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.29
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.15
Experiment series 12
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 533-542
Protein B site: 14-3-3_pThr538
Protein B modification: (TPO)538
Protein B sequence: RHGSPTAPI-
Protein B construct: FRHGSP(pThr)API (crude synthetic peptide)
Average holdup BI: 0.81
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.16
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 4.57
Experiment series 13
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 560-569
Protein B site: 14-3-3_pSer565
Protein B modification: (SEP)565
Protein B sequence: VPRSISVPPS
Protein B construct: VPRSI(pSer)VPPS (crude synthetic peptide)
Average holdup BI: 0.71
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 3.93
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 4.32
Experiment series 14
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1032-1041
Protein B site: 14-3-3_pThr1037
Protein B modification: (TPO)1037
Protein B sequence: IAPYVTLRRG
Protein B construct: IAPYV(pThr)LRRG (crude synthetic peptide)
Average holdup BI: 0.27
Holdup BI standard deviation: 0.09
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.77
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 3.51
Experiment series 15
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 603-612
Protein B site: 14-3-3_pSer608
Protein B modification: (SEP)608
Protein B sequence: RSVDISLGDS
Protein B construct: RSVDI(pSer)LGDS (crude synthetic peptide)
Average holdup BI: -0.06
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.57
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 16
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 603-612
Protein B site: 14-3-3_pSer608
Protein B modification: (SEP)608
Protein B sequence: RSVDISLGDS
Protein B construct: RSVDI(pSer)LGDS (crude synthetic peptide)
Average holdup BI: -0.09
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.53
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 17
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 882-891
Protein B site: 14-3-3_pThr887
Protein B modification: (TPO)887
Protein B sequence: FPQLQTYVPY
Protein B construct: FPQLQ(pThr)YVPY (crude synthetic peptide)
Average holdup BI: 0.31
Holdup BI standard deviation: 0.23
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.4
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 3.59
Experiment series 18
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 897-906
Protein B site: 14-3-3_pThr902
Protein B modification: (TPO)902
Protein B sequence: QLRKVTSPLQ
Protein B construct: QLRKV(pThr)SPLQ (crude synthetic peptide)
Average holdup BI: 0.08
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.34
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 19
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 897-906
Protein B site: 14-3-3_pThr902
Protein B modification: (TPO)902
Protein B sequence: QLRKVTSPLQ
Protein B construct: QLRKV(pThr)SPLQ (crude synthetic peptide)
Average holdup BI: 0.12
Holdup BI standard deviation: 0.1
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.31
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 20
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 599-608
Protein B site: 14-3-3_pSer604
Protein B modification: (SEP)604
Protein B sequence: PDQRRSVDIS
Protein B construct: PDQRR(pSer)VDIS (crude synthetic peptide)
Average holdup BI: 0.07
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.98
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 21
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 599-608
Protein B site: 14-3-3_pSer604
Protein B modification: (SEP)604
Protein B sequence: PDQRRSVDIS
Protein B construct: PDQRR(pSer)VDIS (crude synthetic peptide)
Average holdup BI: 0.06
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.51
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 22
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1105-1114
Protein B site: 14-3-3_pSer1110
Protein B modification: (SEP)1110
Protein B sequence: SSRYLSRPLP
Protein B construct: SSRYL(pSer)RPLP (crude synthetic peptide)
Average holdup BI: 0.07
Holdup BI standard deviation: 0.17
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.29
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 23
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1105-1114
Protein B site: 14-3-3_pSer1110
Protein B modification: (SEP)1110
Protein B sequence: SSRYLSRPLP
Protein B construct: SSRYL(pSer)RPLP (crude synthetic peptide)
Average holdup BI: 0.04
Holdup BI standard deviation: 0.13
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.28
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 24
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1058-1067
Protein B site: 14-3-3_pSer1063
Protein B modification: (SEP)1063
Protein B sequence: LERLYSGDHQ
Protein B construct: LERLY(pSer)GDHQ (crude synthetic peptide)
Average holdup BI: 0.03
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.22
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 25
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 1058-1067
Protein B site: 14-3-3_pSer1063
Protein B modification: (SEP)1063
Protein B sequence: LERLYSGDHQ
Protein B construct: LERLY(pSer)GDHQ (crude synthetic peptide)
Average holdup BI: -0.02
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.17
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 26
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 421-430
Protein B site: 14-3-3_pSer426
Protein B modification: (SEP)426
Protein B sequence: NPEKHSQRKS
Protein B construct: NPEKH(pSer)QRKS (crude synthetic peptide)
Average holdup BI: -0.21
Holdup BI standard deviation: 0.99
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.1
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 27
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: PLEKHA7
Protein B fragment: 443-452
Protein B site: 14-3-3_pSer448
Protein B modification: (SEP)448
Protein B sequence: KGDSRSLPLD
Protein B construct: biotin-ttds-KGDSR(pSer)LPLD (HPLC-purified synthetic peptide)
Measured dissociation constant (10-6M): 93
Standard deviation of dissociation constant (10-6M): 11
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit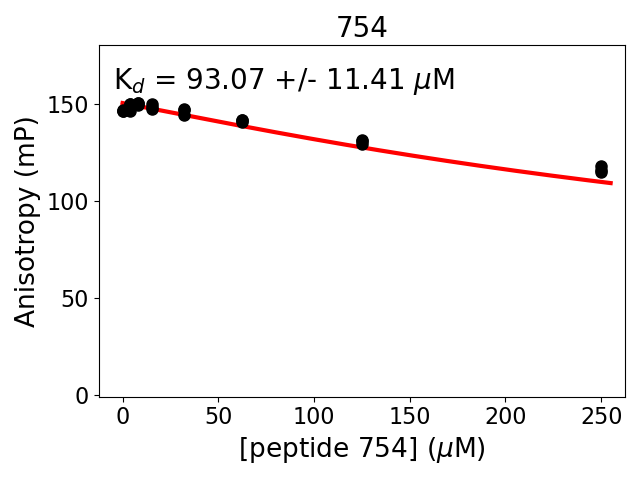
Date of measurement: 2022.02.09
Number of measurements: 3
Measured pKd: 4.03