Details of experiments between P62258 and Q12802
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2406-2415
Protein B site: 14-3-3_pThr2411
Protein B modification: (TPO)2411
Protein B sequence: LFRSNTEEAL
Protein B construct: biotin-ttds-LFRSN(pThr)EEAL (HPLC-purified synthetic peptide)
Measured dissociation constant (10-6M): below threshold
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit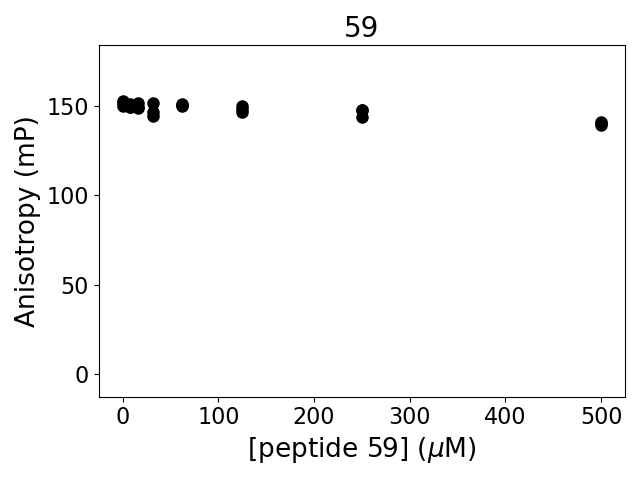
Date of measurement: 2022.02.09
Number of measurements: 3
The affinity was below the detection threshold of the assay.
Experiment series 2
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2462-2471
Protein B site: 14-3-3_pThr2467
Protein B modification: (TPO)2467
Protein B sequence: PRRAETFGGF
Protein B construct: PRRAE(pThr)FGGF (crude synthetic peptide)
Average holdup BI: 0.95
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 8.02
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 5.18
Experiment series 3
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2462-2471
Protein B site: 14-3-3_pThr2467
Protein B modification: (TPO)2467
Protein B sequence: PRRAETFGGF
Protein B construct: PRRAE(pThr)FGGF (crude synthetic peptide)
Average holdup BI: 0.92
Holdup BI standard deviation: 0.01
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.31
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.9
Experiment series 4
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2493-2502
Protein B site: 14-3-3_pSer2498
Protein B modification: (SEP)2498
Protein B sequence: LRRTESDSGL
Protein B construct: LRRTE(pSer)DSGL (crude synthetic peptide)
Average holdup BI: 0.23
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.83
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 5
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1924-1933
Protein B site: 14-3-3_pSer1929
Protein B modification: (SEP)1929
Protein B sequence: KFLSHSTDSL
Protein B construct: KFLSH(pSer)TDSL (crude synthetic peptide)
Average holdup BI: 0.28
Holdup BI standard deviation: 0.07
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 3.75
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.44
Experiment series 6
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1502-1511
Protein B site: 14-3-3_pSer1507
Protein B modification: (SEP)1507
Protein B sequence: SRDRQSLDGF
Protein B construct: SRDRQ(pSer)LDGF (crude synthetic peptide)
Average holdup BI: 0.12
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 3.48
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2493-2502
Protein B site: 14-3-3_pSer2498
Protein B modification: (SEP)2498
Protein B sequence: LRRTESDSGL
Protein B construct: LRRTE(pSer)DSGL (crude synthetic peptide)
Average holdup BI: 0.22
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 3.43
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.29
Experiment series 8
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2406-2415
Protein B site: 14-3-3_pThr2411
Protein B modification: (TPO)2411
Protein B sequence: LFRSNTEEAL
Protein B construct: LFRSN(pThr)EEAL (crude synthetic peptide)
Average holdup BI: 0.09
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 3.3
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1871-1880
Protein B site: 14-3-3_pSer1876
Protein B modification: (SEP)1876
Protein B sequence: KERPRSAVLL
Protein B construct: KERPR(pSer)AVLL (crude synthetic peptide)
Average holdup BI: 0.38
Holdup BI standard deviation: 0.12
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.99
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.63
Experiment series 10
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1637-1646
Protein B site: 14-3-3_pSer1642
Protein B modification: (SEP)1642
Protein B sequence: EERVDSLVSL
Protein B construct: EERVD(pSer)LVSL (crude synthetic peptide)
Average holdup BI: -0.21
Holdup BI standard deviation: 0.1
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.95
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 11
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1871-1880
Protein B site: 14-3-3_pSer1876
Protein B modification: (SEP)1876
Protein B sequence: KERPRSAVLL
Protein B construct: KERPR(pSer)AVLL (crude synthetic peptide)
Average holdup BI: 0.42
Holdup BI standard deviation: 0.14
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.81
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 3.79
Experiment series 12
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1502-1511
Protein B site: 14-3-3_pSer1507
Protein B modification: (SEP)1507
Protein B sequence: SRDRQSLDGF
Protein B construct: SRDRQ(pSer)LDGF (crude synthetic peptide)
Average holdup BI: 0.16
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.81
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.13
Experiment series 13
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2558-2567
Protein B site: 14-3-3_pSer2563
Protein B modification: (SEP)2563
Protein B sequence: LTRSLSRPSS
Protein B construct: LTRSL(pSer)RPSS (crude synthetic peptide)
Average holdup BI: 0.16
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.27
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 14
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1637-1646
Protein B site: 14-3-3_pSer1642
Protein B modification: (SEP)1642
Protein B sequence: EERVDSLVSL
Protein B construct: EERVD(pSer)LVSL (crude synthetic peptide)
Average holdup BI: -0.22
Holdup BI standard deviation: 0.12
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 2.23
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 15
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1924-1933
Protein B site: 14-3-3_pSer1929
Protein B modification: (SEP)1929
Protein B sequence: KFLSHSTDSL
Protein B construct: KFLSH(pSer)TDSL (crude synthetic peptide)
Average holdup BI: 0.16
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.89
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 16
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1640-1649
Protein B site: 14-3-3_pSer1645
Protein B modification: (SEP)1645
Protein B sequence: VDSLVSLSEE
Protein B construct: VDSLV(pSer)LSEE (crude synthetic peptide)
Average holdup BI: -0.24
Holdup BI standard deviation: 0.26
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.44
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 17
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1221-1230
Protein B site: 14-3-3_pSer1226
Protein B modification: (SEP)1226
Protein B sequence: SGRERSTPSL
Protein B construct: SGRER(pSer)TPSL (crude synthetic peptide)
Average holdup BI: 0.09
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.29
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 18
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1745-1754
Protein B site: 14-3-3_pSer1750
Protein B modification: (SEP)1750
Protein B sequence: VSRTFSYIKN
Protein B construct: VSRTF(pSer)YIKN (crude synthetic peptide)
Average holdup BI: 0.14
Holdup BI standard deviation: 0.11
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.29
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 19
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2406-2415
Protein B site: 14-3-3_pThr2411
Protein B modification: (TPO)2411
Protein B sequence: LFRSNTEEAL
Protein B construct: LFRSN(pThr)EEAL (crude synthetic peptide)
Average holdup BI: 0.06
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.23
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 20
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1221-1230
Protein B site: 14-3-3_pSer1226
Protein B modification: (SEP)1226
Protein B sequence: SGRERSTPSL
Protein B construct: SGRER(pSer)TPSL (crude synthetic peptide)
Average holdup BI: 0.05
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.19
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 21
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1745-1754
Protein B site: 14-3-3_pSer1750
Protein B modification: (SEP)1750
Protein B sequence: VSRTFSYIKN
Protein B construct: VSRTF(pSer)YIKN (crude synthetic peptide)
Average holdup BI: 0.09
Holdup BI standard deviation: 0.08
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.15
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 22
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2562-2571
Protein B site: 14-3-3_pSer2567
Protein B modification: (SEP)2567
Protein B sequence: LSRPSSLIEQ
Protein B construct: LSRPS(pSer)LIEQ (crude synthetic peptide)
Average holdup BI: 0.06
Holdup BI standard deviation: 0.07
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.08
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 23
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1640-1649
Protein B site: 14-3-3_pSer1645
Protein B modification: (SEP)1645
Protein B sequence: VDSLVSLSEE
Protein B construct: VDSLV(pSer)LSEE (crude synthetic peptide)
Average holdup BI: 0.15
Holdup BI standard deviation: 0.15
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.06
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 24
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2562-2571
Protein B site: 14-3-3_pSer2567
Protein B modification: (SEP)2567
Protein B sequence: LSRPSSLIEQ
Protein B construct: LSRPS(pSer)LIEQ (crude synthetic peptide)
Average holdup BI: 0.04
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.97
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 25
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1890-1899
Protein B site: 14-3-3_pSer1895
Protein B modification: (SEP)1895
Protein B sequence: FANRRSQQSV
Protein B construct: FANRR(pSer)QQSV (crude synthetic peptide)
Average holdup BI: -0.12
Holdup BI standard deviation: 0.17
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.69
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 26
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1927-1936
Protein B site: 14-3-3_pSer1932
Protein B modification: (SEP)1932
Protein B sequence: SHSTDSLNKI
Protein B construct: SHSTD(pSer)LNKI (crude synthetic peptide)
Average holdup BI: 0.18
Holdup BI standard deviation: 0.28
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.6
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 27
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2468-2477
Protein B site: 14-3-3_pSer2473
Protein B modification: (SEP)2473
Protein B sequence: FGGFDSHQMN
Protein B construct: FGGFD(pSer)HQMN (crude synthetic peptide)
Average holdup BI: 0.19
Holdup BI standard deviation: 0.34
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.47
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 28
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2694-2703
Protein B site: 14-3-3_pSer2699
Protein B modification: (SEP)2699
Protein B sequence: LMRIPSFFPS
Protein B construct: LMRIP(pSer)FFPS (crude synthetic peptide)
Average holdup BI: 0.15
Holdup BI standard deviation: 0.57
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.21
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 29
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 2468-2477
Protein B site: 14-3-3_pSer2473
Protein B modification: (SEP)2473
Protein B sequence: FGGFDSHQMN
Protein B construct: FGGFD(pSer)HQMN (crude synthetic peptide)
Average holdup BI: -0.01
Holdup BI standard deviation: 0.1
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.11
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 30
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1560-1569
Protein B site: 14-3-3_pSer1565
Protein B modification: (SEP)1565
Protein B sequence: PFRRHSWGPG
Protein B construct: PFRRH(pSer)WGPG (crude synthetic peptide)
Average holdup BI: 0.18
Holdup BI standard deviation: 0.83
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.1
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 31
Protein A protein: YWHAE
Protein A fragment: 1-255
Protein A site: 14-3-3
Protein A construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Protein B protein: AKAP13
Protein B fragment: 1890-1899
Protein B site: 14-3-3_pSer1895
Protein B modification: (SEP)1895
Protein B sequence: FANRRSQQSV
Protein B construct: FANRR(pSer)QQSV (crude synthetic peptide)
Average holdup BI: -0.01
Holdup BI standard deviation: 0.27
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.03
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.