Details of experiments between P46937 and P63104
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: YAP1
Protein A fragment: 271-280
Protein A site: 14-3-3_pSer276
Protein A modification: (SEP)276
Protein A sequence: QRISQSAPVK
Protein A construct: QRISQ(pSer)APVK (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.99
Holdup BI standard deviation: 0
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 10.97
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 5.88
Experiment series 2
Protein A protein: YAP1
Protein A fragment: 78-87
Protein A site: 14-3-3_pThr83
Protein A modification: (TPO)83
Protein A sequence: ANVPQTVPMR
Protein A construct: ANVPQ(pThr)VPMR (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.74
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.14
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.34
Experiment series 3
Protein A protein: YAP1
Protein A fragment: 144-153
Protein A site: 14-3-3_pSer149
Protein A modification: (SEP)149
Protein A sequence: PTGVVSGPAA
Protein A construct: PTGVV(pSer)GPAA (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.47
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 5.53
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 3.83
Experiment series 4
Protein A protein: YAP1
Protein A fragment: 366-375
Protein A site: 14-3-3_pSer371
Protein A modification: (SEP)371
Protein A sequence: SSPGMSQELR
Protein A construct: SSPGM(pSer)QELR (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.04
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.86
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 5
Protein A protein: YAP1
Protein A fragment: 376-385
Protein A site: 14-3-3_pSer381
Protein A modification: (SEP)381
Protein A sequence: TMTTNSSDPF
Protein A construct: TMTTN(pSer)SDPF (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.16
Holdup BI standard deviation: 0.13
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.24
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 6
Protein A protein: YAP1
Protein A fragment: 383-392
Protein A site: 14-3-3_pSer388
Protein A modification: (SEP)388
Protein A sequence: DPFLNSGTYH
Protein A construct: DPFLN(pSer)GTYH (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.05
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.92
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: YAP1
Protein A fragment: 89-98
Protein A site: 14-3-3_pSer94
Protein A modification: (SEP)94
Protein A sequence: RKLPDSFFKP
Protein A construct: RKLPD(pSer)FFKP (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.12
Holdup BI standard deviation: 0.54
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.16
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 8
Protein A protein: YAP1
Protein A fragment: 382-391
Protein A site: 14-3-3_pSer387
Protein A modification: (SEP)387
Protein A sequence: TDSGLSMSSY
Protein A construct: TDSGL(pSer)MSSY (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.01
Holdup BI standard deviation: 0.1
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.11
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: YAP1
Protein A fragment: 109-118
Protein A site: 14-3-3_pThr114
Protein A modification: (TPO)114
Protein A sequence: STDAGTAGAL
Protein A construct: STDAG(pThr)AGAL (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0.03
Holdup BI standard deviation: 0.98
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.02
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 10
Protein A protein: YAP1
Protein A fragment: 140-149
Protein A site: 14-3-3_pThr145
Protein A modification: (TPO)145
Protein A sequence: GTLTPTGVVS
Protein A construct: GTLTP(pThr)GVVS (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: 0
Holdup BI standard deviation: 0.09
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.02
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 11
Protein A protein: YAP1
Protein A fragment: 56-65
Protein A site: 14-3-3_pSer61
Protein A modification: (SEP)61
Protein A sequence: HVRGDSETDL
Protein A construct: HVRGD(pSer)ETDL (crude synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Average holdup BI: -0.01
Holdup BI standard deviation: 0.47
Immobilized partner concentration in holdup experiment (10-6M): 132
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.01
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 12
Protein A protein: YAP1
Protein A fragment: 271-280
Protein A site: 14-3-3_pSer276
Protein A modification: (SEP)276
Protein A sequence: QRISQSAPVK
Protein A construct: biotin-ttds-QRISQ(pSer)APVK (HPLC-purified synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): 4.2
Standard deviation of dissociation constant (10-6M): 0.15
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit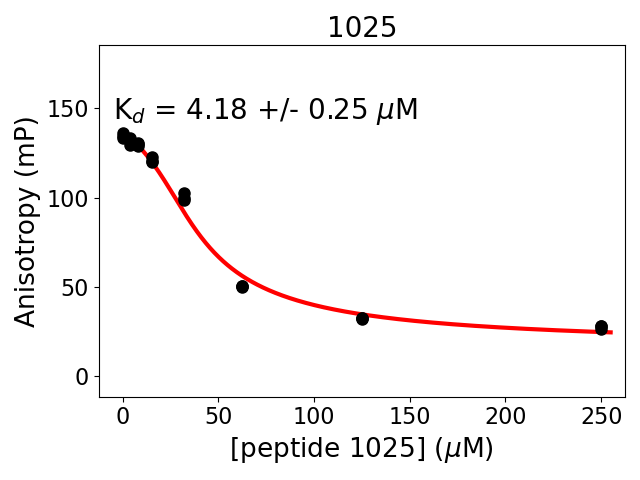
Date of measurement: 2022.01.19
Number of measurements: 3
Measured pKd: 5.38
Experiment series 13
Protein A protein: YAP1
Protein A fragment: 271-280
Protein A site: 14-3-3_pSer276
Protein A modification: (SEP)276
Protein A sequence: QRISQSAPVK
Protein A construct: biotin-ttds-QRISQ(pSer)APVK (HPLC-purified synthetic peptide)
Protein B protein: YWHAZ
Protein B fragment: 1-245
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3zeta
Measured dissociation constant (10-6M): 2.5
Standard deviation of dissociation constant (10-6M): 0.2
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit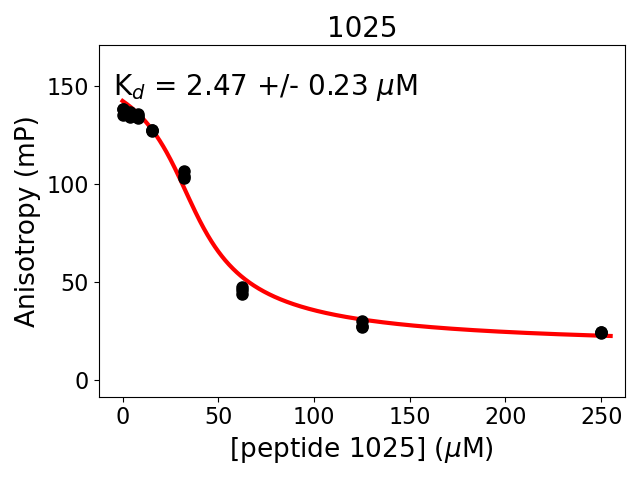
Date of measurement: 2022.02.10
Number of measurements: 3
Measured pKd: 5.61