Details of experiments between O43896 and P62258
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Experiment series 1
Protein A protein: KIF1C
Protein A fragment: 1087-1096
Protein A site: 14-3-3_pSer1092
Protein A modification: (SEP)1092
Protein A sequence: MRRQRSAPDL
Protein A construct: MRRQR(pSer)APDL (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: 0.76
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 7.4
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.34
Experiment series 2
Protein A protein: KIF1C
Protein A fragment: 1077-1086
Protein A site: 14-3-3_pThr1082
Protein A modification: (TPO)1082
Protein A sequence: RYPPYTTPPR
Protein A construct: RYPPY(pThr)TPPR (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: 0.98
Holdup BI standard deviation: 0.02
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 6.52
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 5.55
Experiment series 3
Protein A protein: KIF1C
Protein A fragment: 1087-1096
Protein A site: 14-3-3_pSer1092
Protein A modification: (SEP)1092
Protein A sequence: MRRQRSAPDL
Protein A construct: MRRQR(pSer)APDL (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: 0.71
Holdup BI standard deviation: 0.06
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.86
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
Measured pKd: 4.32
Experiment series 4
Protein A protein: KIF1C
Protein A fragment: 1077-1086
Protein A site: 14-3-3_pThr1082
Protein A modification: (TPO)1082
Protein A sequence: RYPPYTTPPR
Protein A construct: RYPPY(pThr)TPPR (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: 0.89
Holdup BI standard deviation: 0.1
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 4.03
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
Measured pKd: 4.76
Experiment series 5
Protein A protein: KIF1C
Protein A fragment: 484-493
Protein A site: 14-3-3_pThr489
Protein A modification: (TPO)489
Protein A sequence: REDGGTVGVF
Protein A construct: REDGG(pThr)VGVF (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: 0.05
Holdup BI standard deviation: 0.04
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 1.51
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 6
Protein A protein: KIF1C
Protein A fragment: 669-678
Protein A site: 14-3-3_pSer674
Protein A modification: (SEP)674
Protein A sequence: RLYADSDSGD
Protein A construct: RLYAD(pSer)DSGD (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: -0.29
Holdup BI standard deviation: 0.7
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.58
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 7
Protein A protein: KIF1C
Protein A fragment: 272-281
Protein A site: 14-3-3_pThr277
Protein A modification: (TPO)277
Protein A sequence: NKSLTTLGKV
Protein A construct: NKSLT(pThr)LGKV (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: -0.02
Holdup BI standard deviation: 0.05
Immobilized partner concentration in holdup experiment (10-6M): 116
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.32
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2022.02.17.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 4))
Number of measurements: 4
The affinity was below the detection threshold of the assay.
Experiment series 8
Protein A protein: KIF1C
Protein A fragment: 484-493
Protein A site: 14-3-3_pThr489
Protein A modification: (TPO)489
Protein A sequence: REDGGTVGVF
Protein A construct: REDGG(pThr)VGVF (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: 0.01
Holdup BI standard deviation: 0.03
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.26
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 9
Protein A protein: KIF1C
Protein A fragment: 272-281
Protein A site: 14-3-3_pThr277
Protein A modification: (TPO)277
Protein A sequence: NKSLTTLGKV
Protein A construct: NKSLT(pThr)LGKV (crude synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Average holdup BI: -0.03
Holdup BI standard deviation: 0.12
Immobilized partner concentration in holdup experiment (10-6M): 143
P value (usually -log10(P Two-sided unpaired T-test), but double check the experimental details): 0.24
Experiment method: HU Multiplex
Details of affinity fitting: hyperbolic binding equation
Date of measurement: 2021.10.13.
Experimental details: -log10(P Two-sided unpaired T-test (4 vs 5))
Number of measurements: 5
The affinity was below the detection threshold of the assay.
Experiment series 10
Protein A protein: KIF1C
Protein A fragment: 1077-1086
Protein A site: 14-3-3_pThr1082
Protein A modification: (TPO)1082
Protein A sequence: RYPPYTTPPR
Protein A construct: biotin-ttds-RYPPY(pThr)TPPR (HPLC-purified synthetic peptide)
Protein B protein: YWHAE
Protein B fragment: 1-255
Protein B site: 14-3-3
Protein B construct: avi-his6-MBP-TEVsite-14-3-3epsilon
Measured dissociation constant (10-6M): 17
Standard deviation of dissociation constant (10-6M): 3
Experiment method: competitive Fluorescence Polarization
Details of affinity fitting: Competitive binding equation in ProFit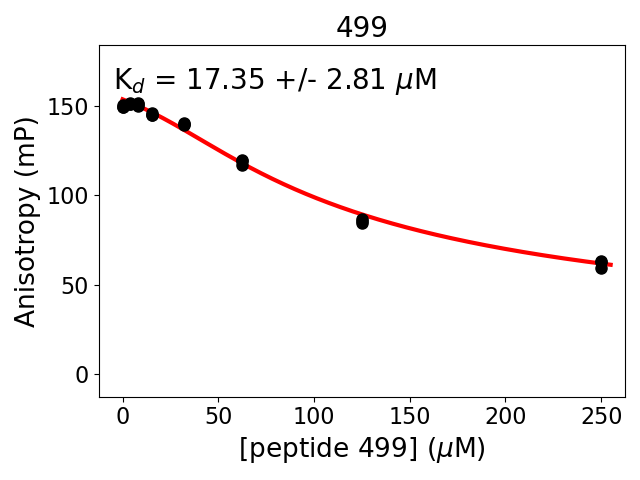
Date of measurement: 2022.02.09
Number of measurements: 3
Measured pKd: 4.76