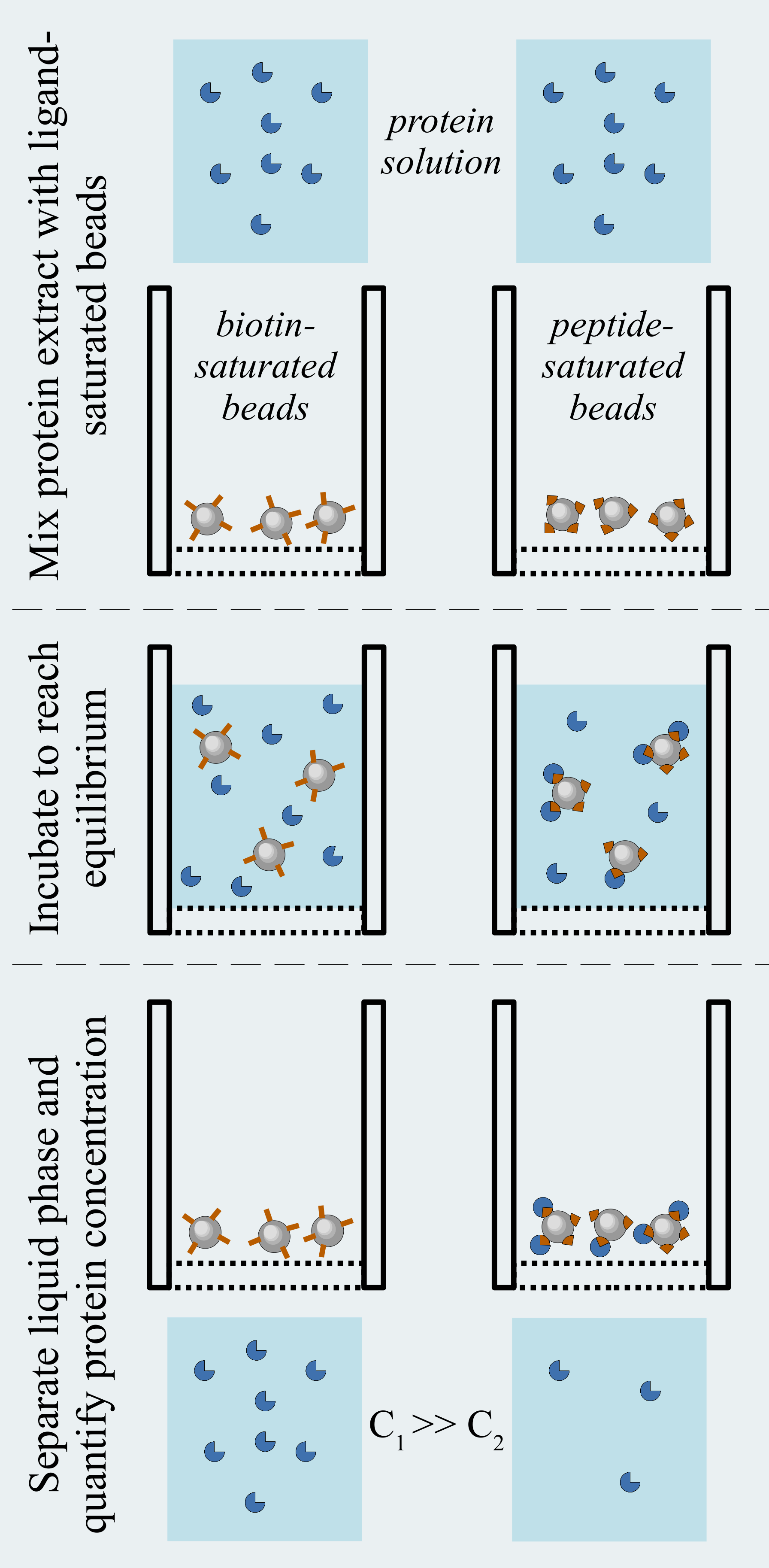
We used the holdup comparative chromatographic retention assay to quantify the affinities of E6-LxxLL interactions. The typical dissociation constants of these complexes are in the micromolar range. In our setup, biotinylated LxxLL peptide motifs were immobilized on streptavidin resin before being incubated with purified MBP-fused E6 proteins in solution. Despite the solubility-enhancing MBP fusion, E6 proteins are prone to self-association. The presence of inactive oligomeric species makes affinity quantification more difficult. This phenomenon is partly due to the presence of non-conserved solvent-exposed cysteine residues, which we mutated to avoid the formation of artifactual intermolecular disulfide bridges. In addition, we systematically used freshly purified MBP-E6 proteins with optimal binding properties for performing the holdup assay.
After incubating MBP-E6 proteins with either LxxLL peptides (ligands) or with biotin-saturated resin (negative control), we recovered by centrifugal filtration the liquid fraction containing the part of MBP-E6 that did not interact with the molecule presented on the resin. By quantifying the depleted protein in both ligand and negative control, we inferred the binding strength of the tested E6-LxxLL interaction. This binding strength is presented by a value called binding intensity, which ranges from 0 (no interaction) to 1 (highest measurable affinity using holdup assay). Similarly to PDZ-PBM interactions, this binding intensity could further be converted into change in Gibbs free energy by estimating the concentration of immobilized ligand on the resin.
Click here for more details.